Development environment
Most useful Packages with python
Data processing
This section list all the packages used for data reading, stats and big data processing.
Pandas
https://pandas.pydata.org/docs/getting_started/intro_tutorials/index.html
Pandas allow a lot of operations :
Read csv files into Dataframes
Resample time series easily
Make statistics
Plot data
conda install -c conda-forge pandas
Xarray
https://xarray.pydata.org/en/stable/
Xarray is a package which handle arrays as Pandas does for the time series :
Open/read/write Netcdf files
Labels array
Easy management of index and dates
Resample, interpolatei
Plot data easily
conda install -c conda-forge xarray
XOA
https://xoa.readthedocs.io/en/latest/index.html
XOA is a package to help reading and manipulating observed and simulated ocean data. It is heavily based on xarray. For those who know it, it is the successor of vacumm.
conda install -c conda-forge xoa
DASK
Dask is a package that allow the user to easily parallelize your data processing, especially when dealing with big data volumes.
conda install dask -c conda-forge
Xskillscore
https://xskillscore.readthedocs.io/en/latest/
Its a package to compute metrics from xarray structures and that return xarray data.
conda install -c conda-forge xskillscore
Plot and figures
Here are the packages used to do all the plots.
Matplotlib
https://matplotlib.org/stable/index.html
The most popular library to make plot and maps
conda install -c conda-forge matplotlib
Cartopy
https://scitools.org.uk/cartopy/docs/latest/gallery/index.html
Cartopy is an extension to Matplotlib dedicated to produce maps from geospatial data
It handle various projections.
conda install -c conda-forge cartopy
Seaborn
Seaborn is a useful package to plot statistical data in Pandas Dataframe format. It’s used here in the part CORA
conda install seaborn -c conda-forge
Python on Datarmor
What tool ?
On datarmor there is a platform to use jupyter notebooks directly on compute nodes
First connect to the following link using the intranet login
https://datarmor-jupyterhub.ifremer.fr
Note
The user can log in once he is connected on the Ifremer network (on the site or via the VPN)
The documentation is available here:
Once connected the user should choose the job profile :
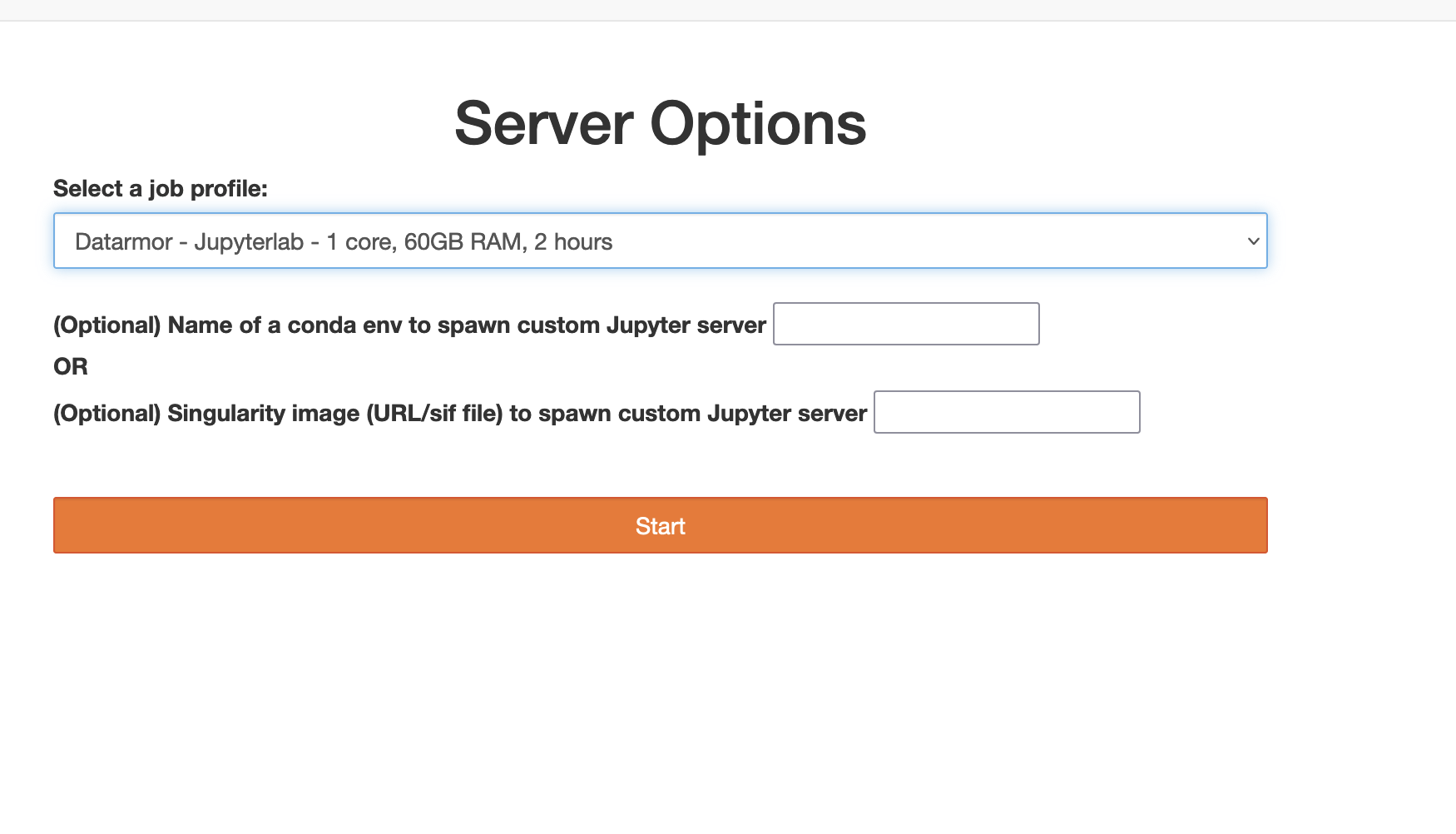
Choose one core if you don’t need to use Dask or if you want to use Dask with distributed job (see next par)
Choose 28 cores if you want to use dask on 28 nodes
Then you have to choose your conda environment
Warning
In order to Jupyterlab to work the user has to install the following packages in the conda environment.
conda install ipykernel jupyterhub jupyterlab nodejs -c conda-forge -y
pip install dask_labextension
Distributed work with Dask on Datarmor
A specification of Dask is, that it’s able to distribute the tasks on different PBS jobs instead of the local one you are connected.
First install the dask-jobqueue package
conda install dask-jobqueue -c conda-forge
To see how it works see the following page:
https://jobqueue.dask.org/en/latest/howitworks.html
So you will sometimes see on top of scripts jobqueue directives for calcul.
The only thing to do is to tell dask-jobqueue the configuration of Datarmor
https://jobqueue.dask.org/en/latest/configuration.html
Here is the configuration file to use and to put here
vi ~/.config/dask/jobqueue.yaml .. code-block:: jobqueue: pbs: name: dask-worker # Dask worker options # number of processes and core have to be equal to avoid using multiple # threads in a single dask worker. Using threads can generate netcdf file # access errors. cores: 28 processes: 6 # this is using all the memory of a single node and corresponds to about # 4GB / dask worker. If you need more memory than this you have to decrease # cores and processes above memory: 120GB interface: ib0 death-timeout: 900 # This should be a local disk attach to your worker node and not a network # mounted disk. See # https://jobqueue.dask.org/en/latest/configuration-setup.html#local-storage # for more details. local-directory: $TMPDIR # PBS resource manager options queue: mpi_1 project: myPROJ walltime: '04:00:00' resource-spec: select=1:ncpus=28:mem=120GB # disable email job-extra: ['-m n'] distributed: scheduler: bandwidth: 1000000000 # GB MB/s estimated worker-worker bandwidth worker: memory: target: 0.90 # Avoid spilling to disk spill: False # Avoid spilling to disk pause: 0.80 # fraction at which we pause worker threads terminate: 0.95 # fraction at which we terminate the worker comm: compression: null